Lab-Made? SARS-CoV-2 Genealogy Through the Lens of Gain-of-Function Research
Yuri Deigin
medium.com

Staff celebrating the physical completion of the laboratory in 2015, Wuhan, China (Source)
If you hear anyone claim “we know the virus didn’t come from a lab”, don’t buy it — it may well have. Labs around the globe have been creating synthetic viruses like CoV2 for years. And no, its genome would not necessarily contain hallmarks of human manipulation: modern genetic engineering tools permit cutting and pasting genomic fragments without leaving a trace. It can be done quickly, too: it took a Swiss team less than a month to create a synthetic clone of CoV2.
How I Learned to Start Worrying
Oh, come on. Lab-made? Nonsense! Back in January, that was my knee-jerk reaction when ideas that Covid-19 is caused by a laboratory leak had just surfaced. Bioweapon? Well, that is just Flat Earth crazies territory. Thus, whenever I kept hearing anything about non-natural origins of SARS-CoV-2, I brushed it aside under similar sentiments. So what if there is a virology institute in Wuhan? Who knows how many of those are sprinkled throughout China.
At some point, it became necessary to brush such theories aside in a substantiated manner, as their proponents began to back up their theses about the possible artificial nature of the virus with arguments from molecular biology, and when engaging them in debate, I wanted to smash their conspiracy theories with cold, hard scientific facts. Just like that Nature paper (or so I thought).
So it was then, in pursuit of arguments against the virus’s lab-madeness, that I got infected by the virus of doubt. What was the source of my doubts? The fact that the deeper you dive into the research activities of coronavirologists over the past 15–20 years, the more you realize that creating chimeras like CoV2 was commonplace in their labs. And CoV2 is an obvious chimera (though not nesessarily a lab-made one), which is based on the ancestral bat strain RaTG13, in which the receptor binding motif (RBM) in its spike protein is replaced by the RBM from a pangolin strain, and in addition, a small but very special stretch of 4 amino acids is inserted, which creates a furin cleavage site that, as virologists have previously established, significantly expands the “repertoire” of the virus in terms of whose cells it can penetrate. Most likely, it was thanks to this new furin site that the new mutant managed to jump species from its original host to humans.
Indeed, virologists, including the leader of coronavirus research at the Wuhan Institute of Virology, Shi Zhengli, have done many similar things in the past — both replacing the RBM in one type of virus by an RBM from another, or adding a new furin site that can provide a species-specific coronavirus with an ability to start using the same receptor (e.g. ACE2) in other species. In fact, Shi Zhengli’s group was creating chimeric constructs as far back as 2007 and as recently as 2017, when they created a whole of 8 new chimeric coronaviruses with various RBMs. In 2019 such work was in full swing, as WIV was part of a $3.7 million NIH grant titled Understanding the Risk of Bat Coronavirus Emergence. Under its auspices, Shi Zhengli co-authored a 2019 paper that called for continued research into synthetic viruses and testing them in vitro and in vivo:
Currently, no clinical treatments or prevention strategies are available for any human coronavirus. Given the conserved RBDs of SARS-CoV and bat SARSr-CoVs, some anti-SARS-CoV strategies in development, such as anti-RBD antibodies or RBD-based vaccines, should be tested against bat SARSr-CoVs. Recent studies demonstrated that anti-SARS-CoV strategies worked against only WIV1 and not SHC014. In addition, little information is available on HKU3-related strains that have much wider geographical distribution and bear truncations in their RBD. Similarly, anti-S antibodies against MERS-CoV could not protect from infection with a pseudovirus bearing the bat MERSr-CoV S. Furthermore, little is known about the replication and pathogenesis of these bat viruses. Thus, future work should be focused on the biological properties of these viruses using virus isolation, reverse genetics and in vitro and in vivo infection assays. The resulting data would help the prevention and control of emerging SARS-like or MERS-like diseases in the future.
If the above quote might seem vague as to what exactly “using reverse genetics” might mean, the NIH grant itself spells it out:
Aim 3. In vitro and in vivo characterization of SARSr-CoV spillover risk, coupled with spatial and phylogenetic analyses to identify the regions and viruses of public health concern. We will use S protein sequence data, infectious clone technology, in vitro and in vivo infection experiments and analysis of receptor binding to test the hypothesis that % divergence thresholds in S protein sequences predict spillover potential.
“Infectious clone technology” stands for creating live synthetic viral clones. Considering the heights of user friendliness and automation that genetic engineering tools have attained, creating a synthetic CoV2 via the above methodology would be in reach of even a grad student.
But before delving into CoV2 origins, let’s first take a quick dive into its biology.
Biology
Ok, let’s start from the basics. What’s a furin site, an RBM, or a spike protein? Bear with me: once you wade through the jungle of terminology, conceptually, everything is pretty straightforward. For example, spike proteins are those red things sticking out of a virus particle — the very reason for which these viruses got “crowned”:
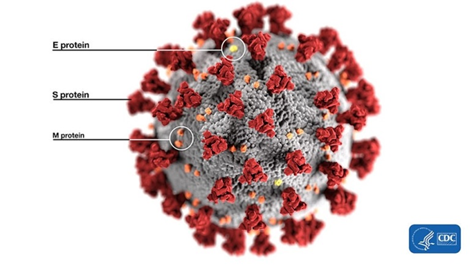
It is with the help of these proteins that the virion clings to the receptor of the victim cell (ACE2 in our case) to then penetrate inside. So it is a vitally important part of the virus, as without getting into a cell viruses cannot replicate. The spike protein also determines which animals the virus can or cannot infect, as ACE2 receptors (or other targets for other viruses) in different species can differ in structure. At the same time, out of the entire 30 kilobase genome (quite huge by viral standards), the gene of this protein makes up only 12–13%. So the spike protein is only about 1300 amino acids long. Below is how the spike (S) protein is structured in CoV2 and close relatives:

As can be seen from the figure above, the S protein consists of two subunits: S1 and S2. It is S1 that interacts with the ACE2 receptor, and the place where S1 does so is called Receptor Binding Domain (RBD), while the area of direct contact, the holy of holies, is called Receptor Binding Motif (RBM). Here is a beautiful illustration from an equally beautiful work:
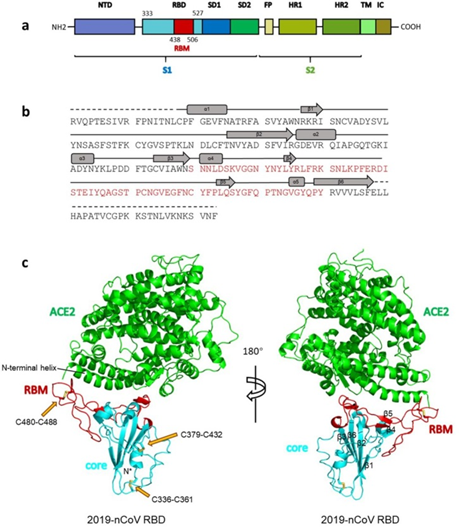
(a) Overall topology of 2019-nCoV spike monomer. NTD, N-terminal domain. RBD, receptor-binding domain. RBM, receptor-binding motif. SD1, subdomain 1. SD2, subdomain 2. FP, fusion peptide. HR1, heptad repeat 1. HR2, heptad repeat 2. TM, transmembrane region. IC, intracellular domain.
(b) Sequence and secondary structures of 2019-nCoV RBD. The RBM is colored red.
© Overall structure of 2019-nCoV RBD bound with ACE2. ACE2 is colored green. 2019-nCoV RBD core is colored cyan and RBM is colored red. Disulfide bonds in the 2019-nCoV RBD are shown as stick and indicated by yellow arrows. The N-terminal helix of ACE2 responsible for binding is labeled.
When the CoV2 genome was just sequenced and made publicly available on January 10, 2020, it was a riddle, as no closely related strains were known. But quite quickly, on January 23, Shi Zhengli released a paper indicating that CoV2 is 96% identical to RaTG13, a strain which her laboratory had previously isolated from Yunnan bats in 2013. However, outside of her lab, no one knew about that strain until January 2020.
It was immediately clear that RaTG13 is special. Take a look at the figure below:
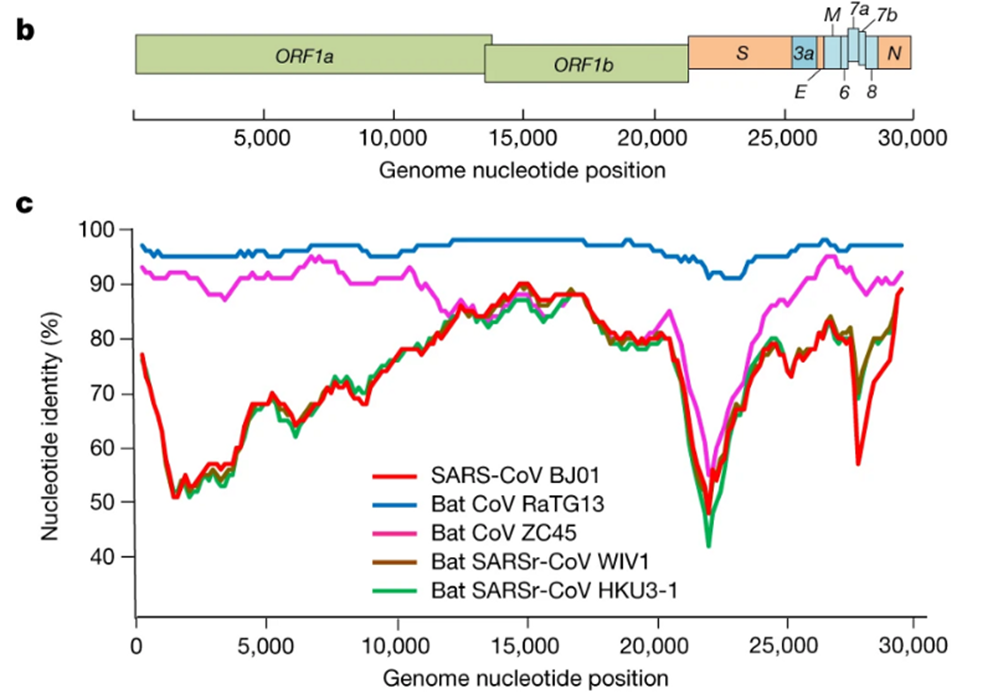
This is a genome similarity graph between CoV2 and other known strains. The higher the curve, the higher the percentage of matching nucleotides. As you can see, in the spike protein (S) gene region (between nucleotides 22k and 25k), only RaTG13 is more or less close to CoV2, while all other strains take a deep dive around this spot — both strains from other bats and the first SARS-CoV (red curve). This in itself is far from suspicious — who knows how many unknown SARS-like strains lurk in the bat caves of Yunnan? Ok, maybe it is not very clear how exactly the virus could get from there to Wuhan, but hey, with those wet markets you never know.
Pangolins
Next, pangolins appeared on the scene: in February, another group of Chinese scientists discovered a peculiar strain of pangolin coronavirus in their possession, which, while generally being only 90% similar to CoV2, in the RBM region was almost identical to it, with only a single amino acid difference (see the upper two sequences, dots indicate a match with the top sequence):
Continue reading at:
https://medium.com/@yurideigin/lab-made-cov2-genealogy-through-the-lens-of-gain-of-function-research-f96dd7413748