Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag
Authors:
Prashant Pradhan
Indian Institute of Technology Delhi
Ashutosh Kumar Pandey
Indian Institute of Technology Delhi
Indian Institute of Technology Delhi
Akhilesh Mishra
Parul Gupta at
Stanford University
Praveen Kumar Tripathi
Indian Institute of Technology Delhi
Manoj Balakrishna Menon
James Gomes
Vivekanandan Perumal at
Indian Institute of Technology Delhi
Bishwajit Kundu
Indian Institute of Technology Delhi
Abstract and Figures
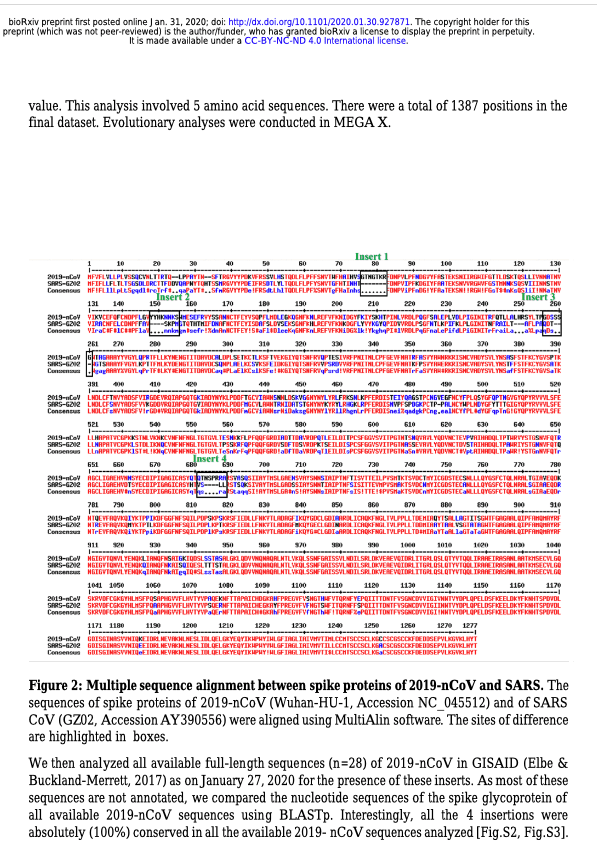
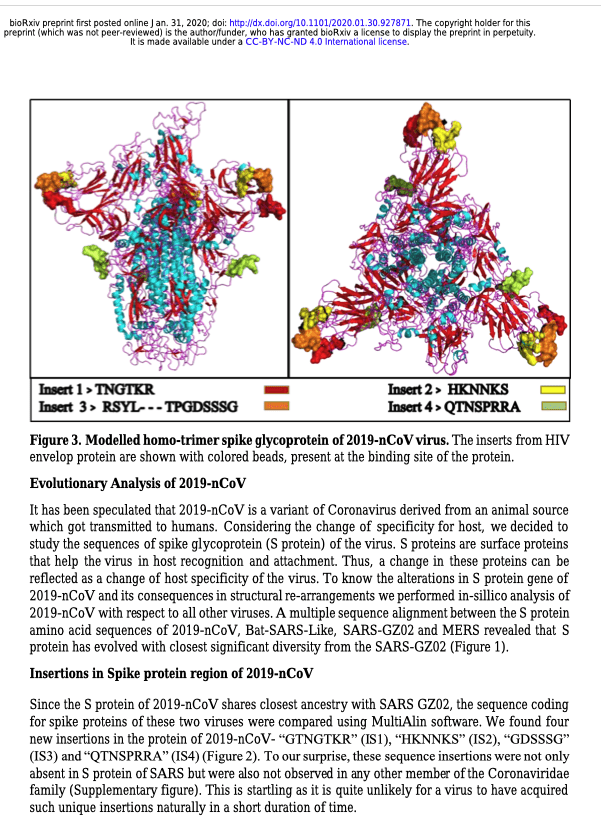
We are currently witnessing a major epidemic caused by the 2019 novel coronavirus (2019- nCoV). The evolution of 2019-nCoV remains elusive. We found 4 insertions in the spike glycoprotein (S) which are unique to the 2019-nCoV and are not present in other coronaviruses. Importantly, amino acid residues in all the 4 inserts have identity or similarity to those in the HIV-1 gp120 or HIV-1 Gag. Interestingly, despite the inserts being discontinuous on the primary amino acid sequence, 3D-modelling of the 2019-nCoV suggests that they converge to constitute the receptor binding site. The finding of 4 unique inserts in the 2019-nCoV, all of which have identity /similarity to amino acid residues in key structural proteins of HIV-1 is unlikely to be fortuitous in nature. This work provides yet unknown insights on 2019-nCoV and sheds light on the evolution and pathogenicity of this virus with important implications for diagnosis of this virus.











Continue reading at:
https://www.researchgate.net/publication/338957445_Uncanny_similarity
_of_unique_inserts_in_the_2019-nCoV_spike_protein_to_HIV-1_gp120_and_Gag